Overview of pathway network databases
Kim Philipp Jablonski, Martin Pirkl
2022-03-31
Source:vignettes/pathway_databases.Rmd
pathway_databases.RmdPathway database overview
We provide access to the following topological pathway databases using graphite (Sales et al. 2012) in a processed format. This format looks as follows:
| database | pathway_id | pathway_name | node_num | edge_num |
|---|---|---|---|---|
| reactome | R-HSA-162582 | Signaling Pathways | 2488 | 62068 |
| reactome | R-HSA-1430728 | Metabolism | 2047 | 85543 |
| reactome | R-HSA-392499 | Metabolism of proteins | 1894 | 52807 |
| reactome | R-HSA-1643685 | Disease | 1774 | 55469 |
| reactome | R-HSA-168256 | Immune System | 1771 | 58277 |
| panther | P00057 | Wnt signaling pathway | 1644 | 195344 |
| reactome | R-HSA-74160 | Gene expression (Transcription) | 1472 | 32493 |
| reactome | R-HSA-597592 | Post-translational protein modification | 1394 | 26399 |
| kegg | hsa:01100 | Metabolic pathways | 1343 | 22504 |
| reactome | R-HSA-73857 | RNA Polymerase II Transcription | 1339 | 25294 |
Let’s see how many pathways each database provides:
dce::df_pathway_statistics %>%
count(database, sort = TRUE, name = "pathway_number") %>%
knitr::kable()| database | pathway_number |
|---|---|
| pathbank | 48685 |
| smpdb | 48671 |
| reactome | 2406 |
| wikipathways | 640 |
| kegg | 323 |
| panther | 94 |
| pharmgkb | 90 |
Next, we can see how the pathway sizes are distributed for each database:
dce::df_pathway_statistics %>%
ggplot(aes(x = node_num)) +
geom_histogram(bins = 30) +
facet_wrap(~ database, scales = "free") +
theme_minimal()
Plotting pathways
It is easily possible to plot pathways:
pathways <- get_pathways(
pathway_list = list(
pathbank = c("Lactose Synthesis"),
kegg = c("Fatty acid biosynthesis")
)
)
lapply(pathways, function(x) {
plot_network(
as(x$graph, "matrix"),
visualize_edge_weights = FALSE,
arrow_size = 0.02,
shadowtext = TRUE
) +
ggtitle(x$pathway_name)
})## [[1]]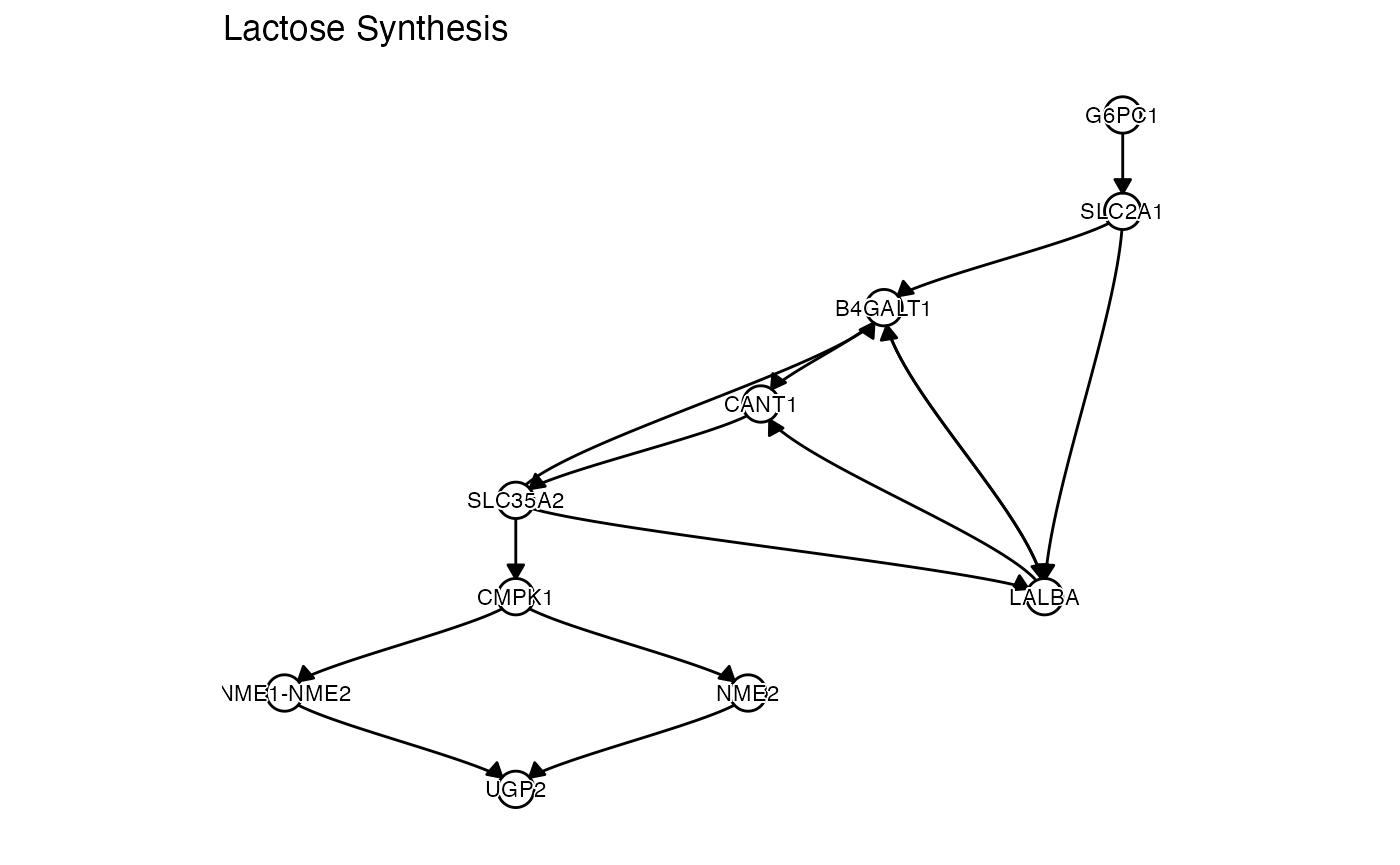
##
## [[2]]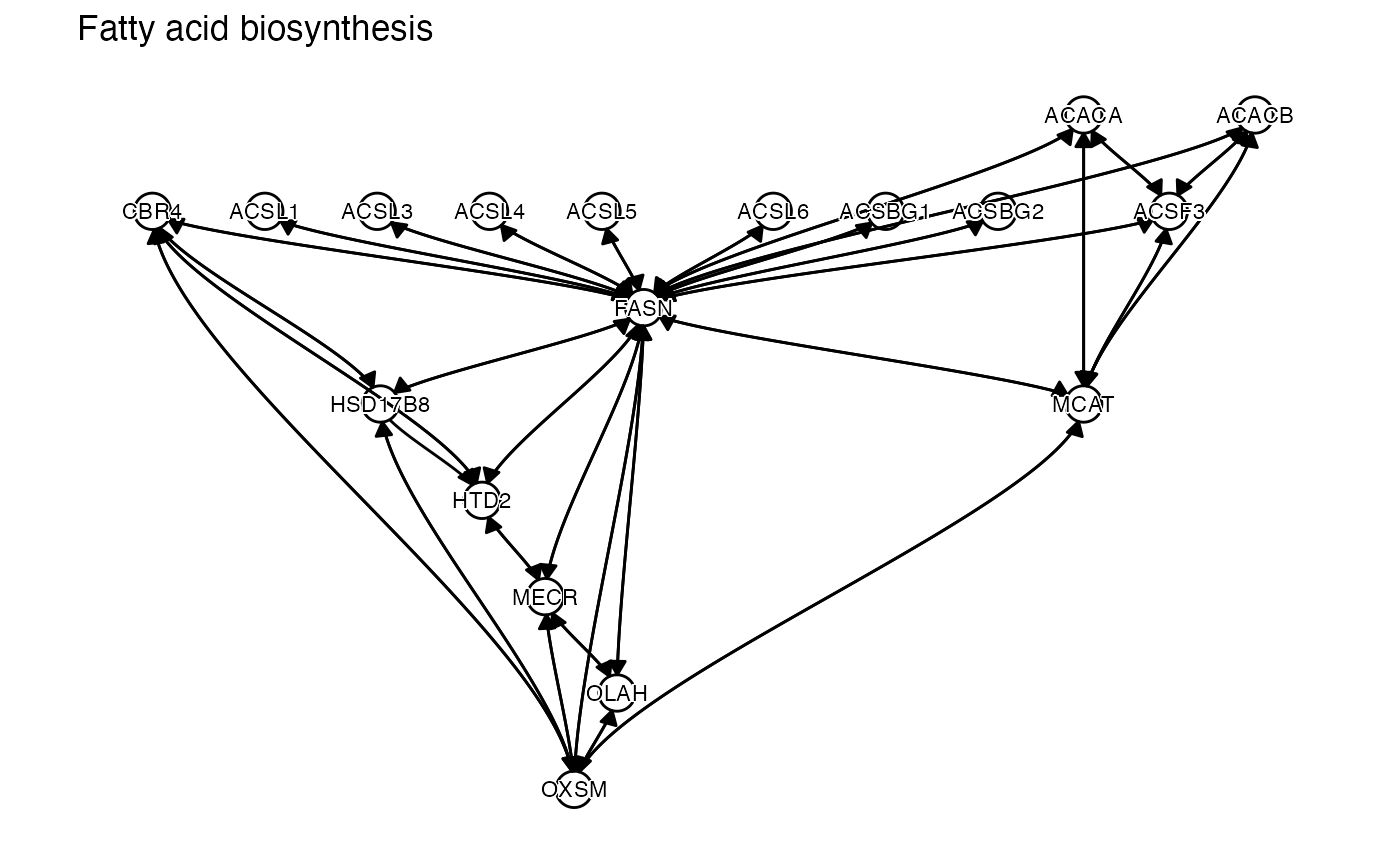
Session information
## R version 4.1.3 (2022-03-10)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur/Monterey 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] org.Hs.eg.db_3.14.0 AnnotationDbi_1.56.2 IRanges_2.28.0
## [4] S4Vectors_0.32.4 Biobase_2.54.0 BiocGenerics_0.40.0
## [7] dce_0.99.6 forcats_0.5.1 stringr_1.4.0
## [10] dplyr_1.0.8 purrr_0.3.4 readr_2.1.2
## [13] tidyr_1.2.0 tibble_3.1.6 ggplot2_3.3.5
## [16] tidyverse_1.3.1 BiocStyle_2.22.0
##
## loaded via a namespace (and not attached):
## [1] rappdirs_0.3.3 prabclus_2.3-2 ragg_1.2.2
## [4] bit64_4.0.5 knitr_1.38 multcomp_1.4-18
## [7] wesanderson_0.3.6 data.table_1.14.2 KEGGREST_1.34.0
## [10] RCurl_1.98-1.6 generics_0.1.2 metap_1.8
## [13] TH.data_1.1-0 RSQLite_2.2.11 shadowtext_0.1.1
## [16] proxy_0.4-26 CombinePValue_1.0 bit_4.0.4
## [19] tzdb_0.3.0 mutoss_0.1-12 xml2_1.3.3
## [22] lubridate_1.8.0 assertthat_0.2.1 viridis_0.6.2
## [25] amap_0.8-18 xfun_0.30 hms_1.1.1
## [28] jquerylib_0.1.4 evaluate_0.15 DEoptimR_1.0-10
## [31] fansi_1.0.3 dbplyr_2.1.1 readxl_1.4.0
## [34] Rgraphviz_2.38.0 igraph_1.2.11 DBI_1.1.2
## [37] tmvnsim_1.0-2 apcluster_1.4.9 RcppArmadillo_0.10.8.1.0
## [40] ellipsis_0.3.2 backports_1.4.1 bookdown_0.25
## [43] permute_0.9-7 harmonicmeanp_3.0 vctrs_0.4.0
## [46] Linnorm_2.18.0 abind_1.4-5 cachem_1.0.6
## [49] RcppEigen_0.3.3.9.1 withr_2.5.0 ggforce_0.3.3
## [52] sfsmisc_1.1-12 robustbase_0.93-9 bdsmatrix_1.3-4
## [55] checkmate_2.0.0 vegan_2.5-7 pcalg_2.7-5
## [58] mclust_5.4.9 mnormt_2.0.2 cluster_2.1.2
## [61] crayon_1.5.1 ellipse_0.4.2 FMStable_0.1-2
## [64] edgeR_3.36.0 pkgconfig_2.0.3 labeling_0.4.2
## [67] qqconf_1.2.3 tweenr_1.0.2 GenomeInfoDb_1.30.1
## [70] nlme_3.1-155 ggm_2.5 nnet_7.3-17
## [73] rlang_1.0.2 diptest_0.76-0 lifecycle_1.0.1
## [76] sandwich_3.0-1 mathjaxr_1.6-0 modelr_0.1.8
## [79] cellranger_1.1.0 rprojroot_2.0.2 polyclip_1.10-0
## [82] matrixStats_0.61.0 graph_1.72.0 Matrix_1.4-0
## [85] zoo_1.8-9 reprex_2.0.1 png_0.1-7
## [88] viridisLite_0.4.0 bitops_1.0-7 Biostrings_2.62.0
## [91] blob_1.2.2 scales_1.1.1 memoise_2.0.1
## [94] graphite_1.40.0 magrittr_2.0.3 plyr_1.8.7
## [97] gdata_2.18.0 zlibbioc_1.40.0 compiler_4.1.3
## [100] plotrix_3.8-2 clue_0.3-60 cli_3.2.0
## [103] XVector_0.34.0 MASS_7.3-55 mgcv_1.8-39
## [106] tidyselect_1.1.2 stringi_1.7.6 textshaping_0.3.6
## [109] highr_0.9 yaml_2.3.5 locfit_1.5-9.5
## [112] ggrepel_0.9.1 grid_4.1.3 sass_0.4.1
## [115] tools_4.1.3 parallel_4.1.3 rstudioapi_0.13
## [118] snowfall_1.84-6.1 gridExtra_2.3 farver_2.1.0
## [121] Rtsne_0.15 ggraph_2.0.5 digest_0.6.29
## [124] BiocManager_1.30.16 flexclust_1.4-0 mnem_1.10.0
## [127] fpc_2.2-9 ppcor_1.1 Rcpp_1.0.8.3
## [130] broom_0.7.12 httr_1.4.2 ggdendro_0.1.23
## [133] kernlab_0.9-29 naturalsort_0.1.3 Rdpack_2.3
## [136] colorspace_2.0-3 rvest_1.0.2 fs_1.5.2
## [139] splines_4.1.3 RBGL_1.70.0 statmod_1.4.36
## [142] sn_2.0.2 expm_0.999-6 pkgdown_2.0.2
## [145] graphlayouts_0.8.0 multtest_2.50.0 flexmix_2.3-17
## [148] systemfonts_1.0.4 jsonlite_1.8.0 tidygraph_1.2.0
## [151] corpcor_1.6.10 modeltools_0.2-23 R6_2.5.1
## [154] gmodels_2.18.1 TFisher_0.2.0 pillar_1.7.0
## [157] htmltools_0.5.2 glue_1.6.2 fastmap_1.1.0
## [160] class_7.3-20 codetools_0.2-18 tsne_0.1-3.1
## [163] mvtnorm_1.1-3 utf8_1.2.2 lattice_0.20-45
## [166] bslib_0.3.1 logger_0.2.2 numDeriv_2016.8-1.1
## [169] curl_4.3.2 gtools_3.9.2 survival_3.2-13
## [172] limma_3.50.1 rmarkdown_2.13 desc_1.4.1
## [175] fastICA_1.2-3 munsell_0.5.0 e1071_1.7-9
## [178] fastcluster_1.2.3 GenomeInfoDbData_1.2.7 haven_2.4.3
## [181] reshape2_1.4.4 gtable_0.3.0 rbibutils_2.2.7